Table of Contents
Worked example:
Data and data source
In this example we use expression data from rat liver 3 hours after vehicle (DMSO) addition.
The file can be downloaded from this link: Data
And was obtained from TG-GATEs project.
Steps:
- Log in PathAct using the login button on the top right corner, the login panel will appear.
- You can also login as anonymous using the start button.
- Upload the file as is shown in the Upload your data section and launch a job with that file.
- A job will appear on the right and will be processed.
- Once finished, click on it to open the view window.
- The view window will appear.
- In this example we simulate the effect of Tamoxifen without using drug assistent. We add manually the targets ESR1 and ESR2 (Estrogen Receptor and Estrogen Receptor Beta) to 0.1, and PRKCA and EBP (Protein kinase C and 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase) to 0.4. Figure 1 shows how add and modify the expression of those genes.
Figure 1: Gene signal modification. Screenshot of gene selection panel. Note when we modify gene activity without drugs, no Additional drug effect appears.
The visualizer highlights modified genes to help locate them (figure 2).
Figure 2: A: Estrogen signaling pathway. Detail of ESR1 modification on the pathway. Note how the viewer marks perturbed genes.
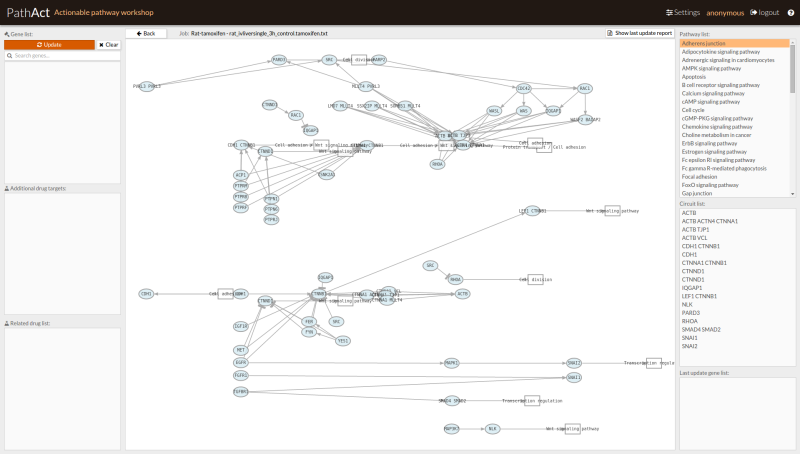
- Perform the inhibition by pressing update button. Those pathways and circuits that have been modified are marked in bold. Red or blue arrows indicate if those changes are or not significant (overactivated path or repressed).
Figure 3: Pathway list aspect after perturbation. The panel marks those pathways (A) and circuits (B) which activation have been modified (bold). Significant modification is marked with red upwards arrows (over activity) and blue down arrows (repression). C: Last gene activity modifications.
Pathways appear painted with red or blue connecting lines in the visualizer. In our example all lines are blue because our inhibition reduce active level of the circuit (figure 4). Figure 5 shows how Estrogen receptors inhibition ends in alteration of transcriptional programs.
Figure 4: A: Thyroid hormone signaling pathway. Significant repressed circuits are painted in blue. B: ESR1 circuit of Thyroid hormone pathway. Note how the circuit selection helps to visualize correctly the perturbed system.
Figure 5: A: Estrogen signaling pathway. Significant repressed circuits are painted in blue. Here we show how In Silico treatment with Tamoxifen decreases Estrogen related paths. B: Detail of Report button. C and D: PathAct report. C: List of perturbed genes and final value of activation. D: ranked circuits by log fold change (base 10). Fold change is used by calculate significance using 2 as threshold (loge 2 = 0.6931472).
- Finally, PathAct reports (figure 5) all the gene signal alterations and circuits log fold change, discriminating between significant or not (absolute FC > 2).